5.3 分类和检测
"医学图像分类和检测正在从计算机辅助检测(CADe)向计算机辅助诊断(CADx)发展,并逐渐成为临床医生的重要助手。" —— AI医学影像发展趋势
在之前的章节中,我们详细学习了预处理技术和基于U-Net的分割方法。现在,我们进入医学图像分析的另一个重要领域:分类和检测。与分割的像素级精度要求不同,分类和检测更关注准确地识别疾病和定位病灶。
医学图像分类和检测面临着独特的挑战:极端的类别不平衡(正负样本比例可达1:1000)、微小的病灶尺寸、图像质量变化,以及对高精度和高召回率的要求。在本节中,我们将探讨如何使用深度学习技术来解决这些问题。
🔍 分类检测核心概念对比
基础任务定义
图像分类
图像分类确定图像是否包含特定的疾病或异常:
- 二分类:正常 vs 异常
- 多类分类:特定疾病类型识别
- 多标签分类:一张图像可能包含多种疾病
目标检测
目标检测不仅识别疾病,还确定其位置:
- 边界框检测:框出病灶区域
- 病灶定位:提供精确坐标
- 多病灶检测:同时检测多个病灶
| 任务类型 | 输入 | 输出 | 临床应用 | 难度等级 |
|---|---|---|---|---|
| 分类 | 完整医学图像 | 疾病标签/类别 | 初筛、分诊 | ⭐⭐ |
| 检测 | 完整医学图像 | 边界框 + 类别 | 病灶定位、手术规划 | ⭐⭐⭐ |
| 分割 | 完整医学图像 | 像素级掩码 | 精确测量、3D重建 | ⭐⭐⭐⭐ |
医学特殊性
类别不平衡问题
医学图像分类面临的核心挑战之一是极端的类别不平衡,这种现象在医学诊断中尤为突出:
🔍 不平衡的根本原因:
疾病流行率差异:正常人群远超患者群体
- 健康体检:95%以上为正常样本
- 疾病筛查:罕见病检出率可能低于0.1%
- 早期诊断:病灶在图像中占比极小
数据采集偏差:
- 医院倾向于收集更多阳性病例用于研究
- 体检中心产生大量正常样本
- 不同科室的病例分布差异巨大
标注成本考虑:
- 阳性样本需要专家详细标注
- 阴性样本相对容易批量标注
- 模糊病例往往被排除在训练集外
⚠️ 不平衡的严重影响:
| 问题类型 | 传统模型表现 | 临床后果 | 解决思路 |
|---|---|---|---|
| 预测偏差 | 倾向预测多数类 | 漏诊高风险疾病 | 加权损失、重采样 |
| 决策阈值 | 固定0.5阈值不适用 | 灵敏度/特异性失衡 | 动态阈值优化 |
| 特征学习 | 忽略少数类特征 | 无法识别病灶特征 | 困难样本挖掘 |
| 评估误导 | 高准确率但无临床价值 | 模型部署风险 | 医学专用指标 |
📊 实际案例分析:
def analyze_class_imbalance(dataset):
"""
分析医学数据集中的类别不平衡
"""
class_counts = {}
for label in dataset.labels:
class_counts[label] = class_counts.get(label, 0) + 1
total_samples = len(dataset)
imbalance_ratios = {}
for class_name, count in class_counts.items():
ratio = count / total_samples
imbalance_ratios[class_name] = {
'count': count,
'percentage': ratio * 100,
'imbalance_factor': max(class_counts.values()) / count
}
return imbalance_ratios
# 示例输出
# {
# 'normal': {'count': 9500, 'percentage': 95.0, 'imbalance_factor': 1.0},
# 'pneumonia': {'count': 300, 'percentage': 3.0, 'imbalance_factor': 31.7},
# 'tuberculosis': {'count': 150, 'percentage': 1.5, 'imbalance_factor': 63.3},
# 'cancer': {'count': 50, 'percentage': 0.5, 'imbalance_factor': 190.0}
# }不确定性和可解释性
医学AI系统的"黑盒"问题是临床应用的主要障碍,医生需要理解模型的决策逻辑才能信任其输出:
🧠 医学决策的特殊要求:
责任归属机制:
- 误诊可能导致严重的法律后果
- 需要明确模型与医生的责任边界
- 可解释性是责任认定的基础
临床决策支持:
- 医生需要验证模型的推理过程
- 理解模型关注的重要特征区域
- 结合临床经验进行综合判断
持续改进需求:
- 通过错误分析改进模型性能
- 识别模型的知识盲区
- 积累罕见病例的处理经验
🔍 可解释性的多层次需求:
| 可解释性层级 | 技术方法 | 临床价值 | 实现复杂度 |
|---|---|---|---|
| 全局解释 | 特征重要性、模型可视化 | 理解模型整体决策逻辑 | ⭐⭐ |
| 局部解释 | 注意力图、Grad-CAM | 解释单次预测的依据 | ⭐⭐⭐ |
| 对比解释 | 反事实解释、原型学习 | 理为什么是A而不是B | ⭐⭐⭐⭐ |
| 不确定性 | 贝叶斯方法、集成学习 | 量化预测的置信度 | ⭐⭐⭐⭐ |
💡 实际应用的可解释性技术:
class MedicalClassificationModel(nn.Module):
"""
可解释的医学分类模型
"""
def __init__(self, backbone, num_classes):
super().__init__()
self.backbone = backbone
self.classifier = nn.Linear(backbone.feature_dim, num_classes)
self.attention_map = nn.Conv2d(backbone.feature_dim, 1, 1)
def forward(self, x):
features = self.backbone.extract_features(x)
# 分类预测
logits = self.classifier(F.adaptive_avg_pool2d(features, 1).flatten(1))
probs = F.softmax(logits, dim=1)
# 注意力图可视化
attention = F.sigmoid(self.attention_map(features))
return {
'predictions': probs,
'attention_maps': attention,
'features': features
}🏥 2D CNN X线图像分类技术
X线分类特征
X线成像的物理基础
X线图像的形成原理决定了其分类任务的独特性:
投影成像特性:
- 三维结构压缩为二维投影
- 不同组织密度产生不同灰度值
- 重叠效应导致信息融合
密度分辨率:
- 骨骼:高密度,白色区域
- 软组织:中等密度,灰色区域
- 气体/脂肪:低密度,黑色区域
临床诊断模式:
- 整体评估:对称性、器官轮廓
- 局部病灶:异常密度、形态改变
- 关联分析:多器官相互影响
整体图像理解的多层次特征
医学影像专家的认知过程反映了深度学习的特征层次:
| 特征层级 | 人类专家认知 | CNN对应层 | 临床意义 |
|---|---|---|---|
| 低级特征 | 边缘、纹理、对比度 | 浅层卷积 | 器官边界、密度差异 |
| 中级特征 | 形状模式、结构关系 | 中层卷积 | 器官形态、相对位置 |
| 高级特征 | 病理征象、诊断模式 | 深层全连接 | 疾病模式、预后判断 |
🏥 不同解剖部位的X线分类特点:
胸部X线(最常见的分类任务):
- 全局对称性:左右肺野的对称性评估
- 纵隔位置:气管、心脏位置偏移
- 膈肌形态:膈面光滑度、位置高度
- 肺纹理分布:血管纹理的粗细、分布
骨关节X线:
- 骨骼完整性:骨折、骨质破坏
- 关节间隙:关节炎、关节退变
- 软组织肿胀:炎症、外伤征象
腹部X线:
- 肠道气体分布:肠梗阻、穿孔
- 钙化灶识别:结石、结核钙化
- 脏器轮廓:肝脾增大、腹水征象
🔍 分类任务的决策路径:
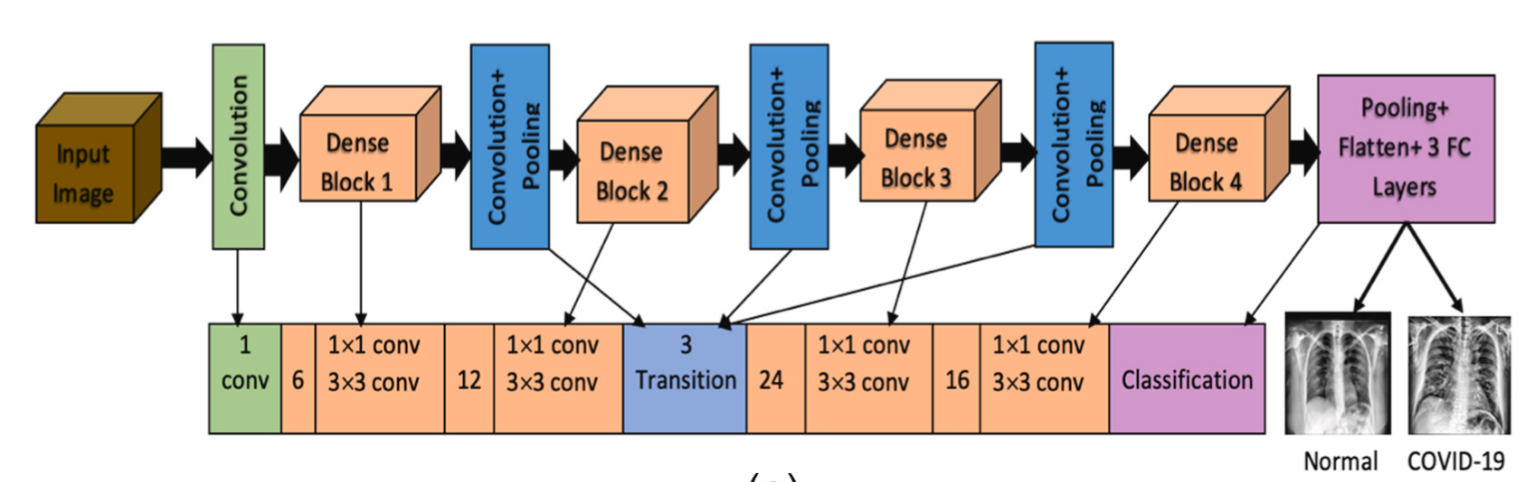 基于深度学习的胸部X线分类,模型自动学习疾病特征Source: Diagnosis of corona diseases from associated genes and X-ray images using machine learning algorithms and deep CNN - 基于机器学习算法和深度CNN使用关联基因和X光图像诊断冠状病毒疾病
基于深度学习的胸部X线分类,模型自动学习疾病特征Source: Diagnosis of corona diseases from associated genes and X-ray images using machine learning algorithms and deep CNN - 基于机器学习算法和深度CNN使用关联基因和X光图像诊断冠状病毒疾病
经典2D CNN架构
基于ResNet的医学分类
import torch.nn as nn
import torch.nn.functional as F
from torchvision.models import resnet50
class MedicalResNet(nn.Module):
"""
基于ResNet的医学图像分类模型
"""
def __init__(self, num_classes, pretrained=True, dropout_rate=0.5):
super().__init__()
# 加载预训练ResNet
self.backbone = resnet50(pretrained=pretrained)
# 修改第一层以适应医学灰度图像
self.backbone.conv1 = nn.Conv2d(
1, 64, kernel_size=7, stride=2, padding=3, bias=False
)
# 替换分类器
feature_dim = self.backbone.fc.in_features
self.backbone.fc = nn.Identity()
# 自定义分类头
self.classifier = nn.Sequential(
nn.AdaptiveAvgPool2d(1),
nn.Flatten(),
nn.Dropout(dropout_rate),
nn.Linear(feature_dim, 512),
nn.ReLU(inplace=True),
nn.Dropout(dropout_rate * 0.5),
nn.Linear(512, num_classes)
)
def forward(self, x):
features = self.backbone(x)
return self.classifier(features)DenseNet的优势
DenseNet特别适合医学图像分类:
class DenseNetMedical(nn.Module):
"""
基于DenseNet的医学分类模型
"""
def __init__(self, num_classes, growth_rate=32, block_config=(6, 12, 24, 16)):
super().__init__()
# DenseNet特征提取器
self.features = self._make_dense_layers(growth_rate, block_config)
# 分类头
self.classifier = nn.Sequential(
nn.BatchNorm2d(block_config[-1] * growth_rate),
nn.ReLU(inplace=True),
nn.AdaptiveAvgPool2d(1),
nn.Flatten(),
nn.Linear(block_config[-1] * growth_rate, num_classes)
)
def _make_dense_layers(self, growth_rate, block_config):
"""
构建DenseNet层
"""
layers = []
# 初始卷积
layers += [
nn.Conv2d(1, 2 * growth_rate, 7, stride=2, padding=3, bias=False),
nn.BatchNorm2d(2 * growth_rate),
nn.ReLU(inplace=True),
nn.MaxPool2d(3, stride=2, padding=1)
]
# 密集块
num_features = 2 * growth_rate
for i, num_layers in enumerate(block_config):
layers.append(self._make_dense_block(num_features, num_layers, growth_rate))
num_features += num_layers * growth_rate
if i != len(block_config) - 1:
layers.append(self._make_transition_layer(num_features, num_features // 2))
num_features = num_features // 2
return nn.Sequential(*layers)
def _make_dense_block(self, in_channels, num_layers, growth_rate):
"""
构建密集块
"""
layers = []
for _ in range(num_layers):
layers.append(self._make_dense_layer(in_channels, growth_rate))
in_channels += growth_rate
return nn.Sequential(*layers)
def _make_dense_layer(self, in_channels, growth_rate):
"""
构建密集层
"""
return nn.Sequential(
nn.BatchNorm2d(in_channels),
nn.ReLU(inplace=True),
nn.Conv2d(in_channels, 4 * growth_rate, 1, bias=False),
nn.BatchNorm2d(4 * growth_rate),
nn.ReLU(inplace=True),
nn.Conv2d(4 * growth_rate, growth_rate, 3, padding=1, bias=False)
)数据不平衡处理策略
损失函数设计
class FocalLoss(nn.Module):
"""
Focal Loss:解决类别不平衡问题
"""
def __init__(self, alpha=1, gamma=2, reduction='mean'):
super().__init__()
self.alpha = alpha
self.gamma = gamma
self.reduction = reduction
def forward(self, inputs, targets):
ce_loss = F.cross_entropy(inputs, targets, reduction='none')
pt = torch.exp(-ce_loss)
focal_loss = self.alpha * (1 - pt) ** self.gamma * ce_loss
if self.reduction == 'mean':
return focal_loss.mean()
elif self.reduction == 'sum':
return focal_loss.sum()
else:
return focal_loss
class BalancedCrossEntropyLoss(nn.Module):
"""
平衡交叉熵损失
"""
def __init__(self, class_weights=None):
super().__init__()
self.class_weights = class_weights
def forward(self, inputs, targets):
if self.class_weights is not None:
weight = self.class_weights[targets]
ce_loss = F.cross_entropy(inputs, targets, weight=weight, reduction='none')
else:
ce_loss = F.cross_entropy(inputs, targets, reduction='none')
return ce_loss.mean()数据采样策略
class BalancedSampler(torch.utils.data.Sampler):
"""
平衡采样器:确保每个batch有均衡的样本
"""
def __init__(self, dataset, samples_per_class):
self.dataset = dataset
self.samples_per_class = samples_per_class
# 按类别分组
self.class_indices = {}
for idx, label in enumerate(dataset.labels):
if label not in self.class_indices:
self.class_indices[label] = []
self.class_indices[label].append(idx)
def __iter__(self):
batch = []
for _ in range(len(self) // self.samples_per_class):
# 从每个类别随机选择样本
sampled_indices = []
for class_indices in self.class_indices.values():
if len(class_indices) > 0:
sampled_idx = np.random.choice(class_indices)
sampled_indices.append(sampled_idx)
batch.extend(sampled_indices)
np.random.shuffle(batch)
return iter(batch)
def __len__(self):
return len(self.dataset)🧠 3D CNN体积数据分析
从2D到3D:挑战与机遇
3D数据特征
体积医学数据(CT、MRI)具有独特特征:
| 特征 | 2D图像 | 3D体积 |
|---|---|---|
| 空间信息 | 平面上下文 | 完整3D空间关系 |
| 计算复杂度 | O(H×W) | O(H×W×D) |
| 内存需求 | MB | GB |
| 层间信息 | 丢失 | 保留 |
| 临床价值 | 局部诊断 | 综合分析 |
3D卷积操作
class Conv3DBlock(nn.Module):
"""
3D卷积块
"""
def __init__(self, in_channels, out_channels, kernel_size=3, stride=1, padding=1):
super().__init__()
self.conv = nn.Conv3d(in_channels, out_channels, kernel_size, stride, padding, bias=False)
self.bn = nn.BatchNorm3d(out_channels)
self.relu = nn.ReLU(inplace=True)
def forward(self, x):
return self.relu(self.bn(self.conv(x)))
class ResNet3DBlock(nn.Module):
"""
3D残差块
"""
def __init__(self, in_channels, out_channels, stride=1):
super().__init__()
self.conv1 = Conv3DBlock(in_channels, out_channels, stride=stride)
self.conv2 = nn.Sequential(
nn.Conv3d(out_channels, out_channels, 3, padding=1, bias=False),
nn.BatchNorm3d(out_channels)
)
self.shortcut = nn.Sequential()
if stride != 1 or in_channels != out_channels:
self.shortcut = nn.Sequential(
nn.Conv3d(in_channels, out_channels, 1, stride, bias=False),
nn.BatchNorm3d(out_channels)
)
def forward(self, x):
residual = self.shortcut(x)
out = self.conv1(x)
out = self.conv2(out)
out += residual
return F.relu(out)3D CNN架构设计
医学3D ResNet
class Medical3DResNet(nn.Module):
"""
用于体积医学图像分类的3D ResNet
"""
def __init__(self, block, layers, num_classes=2):
super().__init__()
self.in_channels = 64
# 初始卷积
self.conv1 = nn.Sequential(
nn.Conv3d(1, 64, kernel_size=7, stride=2, padding=3, bias=False),
nn.BatchNorm3d(64),
nn.ReLU(inplace=True),
nn.MaxPool3d(kernel_size=3, stride=2, padding=1)
)
# 残差块
self.layer1 = self._make_layer(block, 64, layers[0])
self.layer2 = self._make_layer(block, 128, layers[1], stride=2)
self.layer3 = self._make_layer(block, 256, layers[2], stride=2)
self.layer4 = self._make_layer(block, 512, layers[3], stride=2)
# 分类头
self.avgpool = nn.AdaptiveAvgPool3d((1, 1, 1))
self.fc = nn.Linear(512 * block.expansion, num_classes)
# 初始化权重
self._initialize_weights()
def _make_layer(self, block, out_channels, blocks, stride=1):
layers = []
layers.append(block(self.in_channels, out_channels, stride))
self.in_channels = out_channels * block.expansion
for _ in range(1, blocks):
layers.append(block(self.in_channels, out_channels))
return nn.Sequential(*layers)
def _initialize_weights(self):
for m in self.modules():
if isinstance(m, nn.Conv3d):
nn.init.kaiming_normal_(m.weight, mode='fan_out', nonlinearity='relu')
elif isinstance(m, nn.BatchNorm3d):
nn.init.constant_(m.weight, 1)
nn.init.constant_(m.bias, 0)
def forward(self, x):
x = self.conv1(x)
x = self.layer1(x)
x = self.layer2(x)
x = self.layer3(x)
x = self.layer4(x)
x = self.avgpool(x)
x = torch.flatten(x, 1)
x = self.fc(x)
return x
# 模型实例化
def create_medical_3d_resnet(num_classes=2):
return Medical3DResNet(ResNet3DBlock, [2, 2, 2, 2], num_classes)3D DenseNet架构
class DenseBlock3D(nn.Module):
"""
3D密集块
"""
def __init__(self, num_layers, in_channels, growth_rate):
super().__init__()
self.layers = nn.ModuleList()
for i in range(num_layers):
layer = nn.Sequential(
nn.BatchNorm3d(in_channels + i * growth_rate),
nn.ReLU(inplace=True),
nn.Conv3d(in_channels + i * growth_rate, 4 * growth_rate, 1, bias=False),
nn.BatchNorm3d(4 * growth_rate),
nn.ReLU(inplace=True),
nn.Conv3d(4 * growth_rate, growth_rate, 3, padding=1, bias=False)
)
self.layers.append(layer)
def forward(self, x):
features = [x]
for layer in self.layers:
new_features = layer(torch.cat(features, dim=1))
features.append(new_features)
return torch.cat(features, dim=1)内存优化策略
基于Patches的训练
class Patch3DDataset(torch.utils.data.Dataset):
"""
3D patch数据集
"""
def __init__(self, volume_path, label_path, patch_size=(64, 64, 64), stride=32):
self.patch_size = patch_size
self.stride = stride
# 加载体积和标签
self.volume = nib.load(volume_path).get_fdata()
self.label = nib.load(label_path).get_fdata()
# 生成patch坐标
self.patches = self._generate_patches()
def _generate_patches(self):
"""
生成3D patch坐标
"""
patches = []
D, H, W = self.volume.shape
pd, ph, pw = self.patch_size
for d in range(0, D - pd + 1, self.stride):
for h in range(0, H - ph + 1, self.stride):
for w in range(0, W - pw + 1, self.stride):
patch_volume = self.volume[d:d+pd, h:h+ph, w:w+pw]
patch_label = self.label[d:d+pd, h:h+ph, w:w+pw]
# 过滤掉背景patches
if np.sum(patch_label > 0) > 0.1 * np.prod(self.patch_size):
patches.append((d, h, w))
return patches
def __getitem__(self, idx):
d, h, w = self.patches[idx]
pd, ph, pw = self.patch_size
volume_patch = self.volume[d:d+pd, h:h+ph, w:w+pw]
label_patch = self.label[d:d+pd, h:h+ph, w:w+pw]
# 归一化
volume_patch = self._normalize_volume(volume_patch)
return torch.FloatTensor(volume_patch).unsqueeze(0), torch.LongTensor([label_patch.max()])梯度检查点
from torch.utils.checkpoint import checkpoint
class MemoryEfficient3DNet(nn.Module):
"""
内存高效的3D网络
"""
def __init__(self):
super().__init__()
self.layer1 = self._create_layer(1, 64)
self.layer2 = self._create_layer(64, 128)
self.layer3 = self._create_layer(128, 256)
self.layer4 = self._create_layer(256, 512)
def _create_layer(self, in_channels, out_channels):
return nn.Sequential(
nn.Conv3d(in_channels, out_channels, 3, padding=1),
nn.BatchNorm3d(out_channels),
nn.ReLU(inplace=True),
nn.Conv3d(out_channels, out_channels, 3, padding=1),
nn.BatchNorm3d(out_channels),
nn.ReLU(inplace=True)
)
def forward(self, x):
# 使用梯度检查点节省内存
x1 = checkpoint(self.layer1, x)
x2 = checkpoint(self.layer2, x1)
x3 = checkpoint(self.layer3, x2)
x4 = checkpoint(self.layer4, x3)
return x4🔬 全幻灯片图像多示例学习
全幻灯片图像挑战
WSI特征
全幻灯片图像(WSI)具有独特特征:
- 超高分辨率:100,000×100,000像素或更高
- 巨大文件大小:数GB
- 可变放大倍数:不同物镜(4x、10x、20x、40x)
- 仅有幻灯片级标签:通常只知道患者是否患有癌症
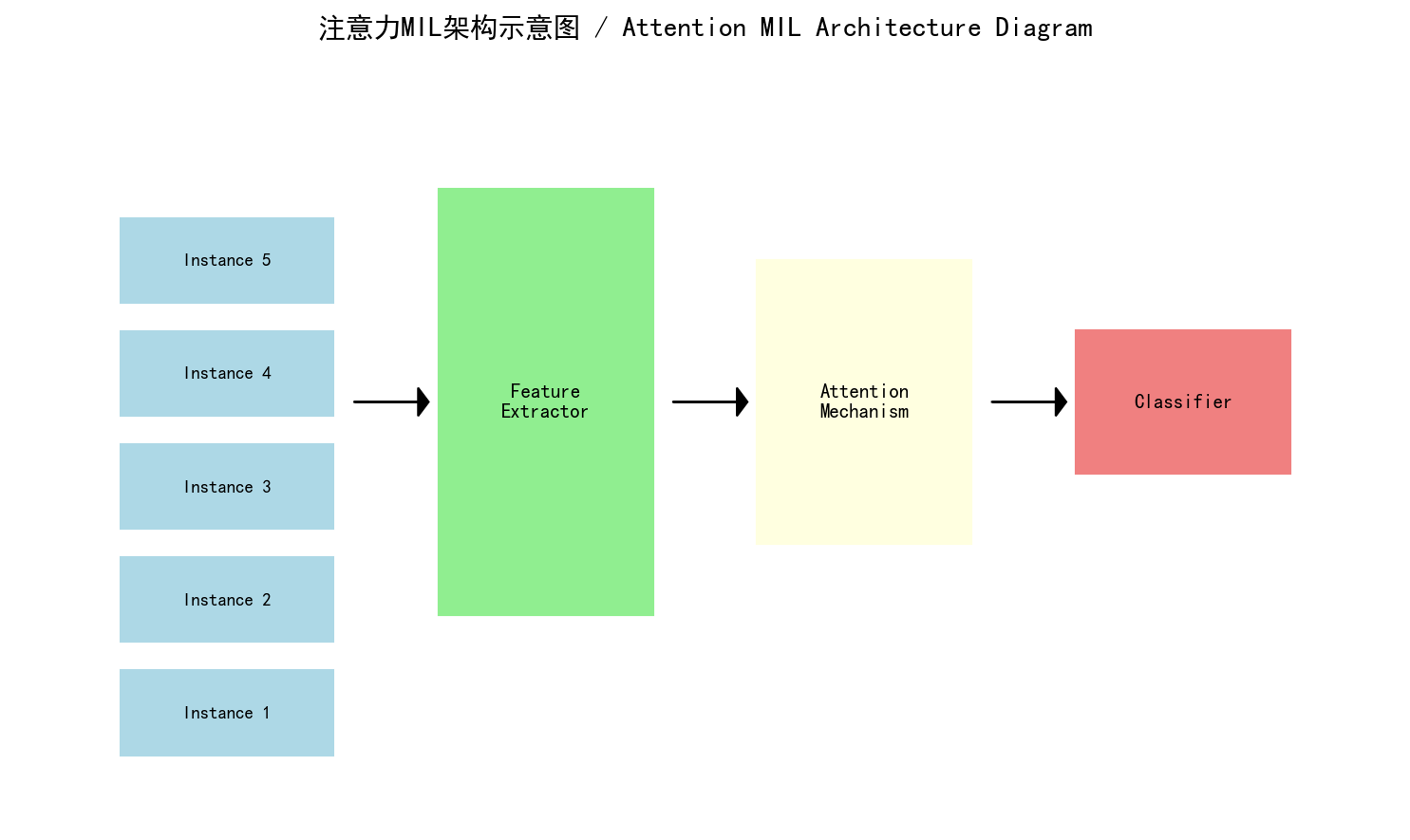 基于MIL的全幻灯片图像分析工作流程 - 自制示意图,展示从WSI到最终诊断的流程
基于MIL的全幻灯片图像分析工作流程 - 自制示意图,展示从WSI到最终诊断的流程
MIL基础
问题形式化
在MIL中,一张幻灯片被视为一个包(bag),从中提取的patches是实例(instances):
- 包标签:已知(如癌性/正常)
- 实例标签:未知(哪些patches包含癌症)
- 假设:如果包是阳性的,至少有一个实例是阳性的
class MILDataset(torch.utils.data.Dataset):
"""
多示例学习数据集
"""
def __init__(self, wsi_paths, labels, patch_size=256, magnification=20):
self.wsi_paths = wsi_paths
self.labels = labels
self.patch_size = patch_size
self.magnification = magnification
# 预提取patches或动态提取
self.patch_cache = {}
def extract_patches(self, wsi_path, num_patches=100):
"""
从WSI提取patches
"""
wsi = openslide.OpenSlide(wsi_path)
# 获取图像尺寸
level = self._get_best_level(wsi, self.magnification)
dimensions = wsi.level_dimensions[level]
patches = []
for _ in range(num_patches):
# 随机采样
x = np.random.randint(0, dimensions[0] - self.patch_size)
y = np.random.randint(0, dimensions[1] - self.patch_size)
patch = wsi.read_region((x, y), level,
(self.patch_size, self.patch_size))
patch = patch.convert('RGB')
# 组织过滤
if self._is_tissue_patch(patch):
patches.append(np.array(patch))
return patches
def _is_tissue_patch(self, patch, threshold=0.1):
"""
检查patch是否包含组织
"""
# 转换到HSV空间进行组织检测
hsv = cv2.cvtColor(patch, cv2.COLOR_RGB2HSV)
# 组织通常出现在特定的H、S范围内
tissue_mask = (hsv[:, :, 0] > 0.1) & (hsv[:, :, 1] > 0.1)
tissue_ratio = np.sum(tissue_mask) / patch.size
return tissue_ratio > threshold
def __getitem__(self, idx):
if idx not in self.patch_cache:
self.patch_cache[idx] = self.extract_patches(self.wsi_paths[idx])
patches = self.patch_cache[idx]
label = self.labels[idx]
# 转换为tensor
patches_tensor = torch.FloatTensor(np.array(patches)).permute(0, 3, 1, 2)
label_tensor = torch.LongTensor([label])
return patches_tensor, label_tensorMIL算法实现
实例级注意力
class AttentionMIL(nn.Module):
"""
基于注意力的多示例学习模型
"""
def __init__(self, input_size=256*256*3, hidden_size=512, attention_size=256, num_classes=2):
super().__init__()
# 特征提取器
self.feature_extractor = nn.Sequential(
nn.Linear(input_size, hidden_size),
nn.ReLU(),
nn.Dropout(0.5),
nn.Linear(hidden_size, hidden_size),
nn.ReLU(),
nn.Dropout(0.5)
)
# 注意力机制
self.attention = nn.Sequential(
nn.Linear(hidden_size, attention_size),
nn.Tanh(),
nn.Linear(attention_size, 1)
)
# 分类器
self.classifier = nn.Linear(hidden_size, num_classes)
def forward(self, bag):
"""
前向传播
Args:
bag: (batch_size, num_instances, channels, height, width)
"""
batch_size, num_instances, C, H, W = bag.shape
# 重塑以处理每个实例
bag = bag.view(-1, C, H, W)
# 特征提取
features = self.feature_extractor(bag.view(bag.size(0), -1))
# 重塑回bag格式
features = features.view(batch_size, num_instances, -1)
# 注意力计算
attention_weights = self.attention(features) # (batch_size, num_instances, 1)
attention_weights = F.softmax(attention_weights, dim=1)
# 加权池化
weighted_features = torch.sum(features * attention_weights, dim=1)
# 分类
logits = self.classifier(weighted_features)
return {
'logits': logits,
'attention_weights': attention_weights,
'instance_features': features
}
def calculate_loss(self, outputs, targets, attention_reg_strength=0.001):
"""
计算MIL损失
"""
# 分类损失
classification_loss = F.cross_entropy(outputs['logits'], targets)
# 注意力正则化(防止注意力只集中在单个实例上)
attention_entropy = -torch.mean(
torch.sum(outputs['attention_weights'] *
torch.log(outputs['attention_weights'] + 1e-8), dim=1)
)
total_loss = classification_loss - attention_reg_strength * attention_entropy
return total_loss深度MIL框架
class DeepMIL(nn.Module):
"""
深度多示例学习框架
"""
def __init__(self, backbone='resnet50', mil_type='attention', num_classes=2):
super().__init__()
self.backbone = self._create_backbone(backbone)
self.mil_layer = self._create_mil_layer(mil_type)
self.classifier = nn.Linear(2048, num_classes) # ResNet50输出维度
def _create_backbone(self, backbone_name):
"""
创建CNN backbone
"""
if backbone_name == 'resnet50':
from torchvision.models import resnet50
backbone = resnet50(pretrained=True)
# 移除最后的全连接层
backbone = nn.Sequential(*list(backbone.children())[:-1])
return backbone
def _create_mil_layer(self, mil_type):
"""
创建MIL层
"""
if mil_type == 'attention':
return AttentionMIL()
elif mil_type == 'max_pooling':
return nn.AdaptiveMaxPool2d(1)
elif mil_type == 'mean_pooling':
return nn.AdaptiveAvgPool2d(1)
else:
raise ValueError(f"不支持的MIL类型: {mil_type}")
def forward(self, bag):
"""
前向传播
Args:
bag: (batch_size, num_instances, 3, H, W)
"""
batch_size, num_instances = bag.shape[:2]
# 重塑以处理每个实例
instances = bag.view(-1, *bag.shape[2:])
# 特征提取
with torch.no_grad():
features = self.backbone(instances) # (batch_size * num_instances, 2048, 1, 1)
features = features.view(features.size(0), -1) # 展平
# 重塑为bag格式
features = features.view(batch_size, num_instances, -1)
# MIL聚合
if isinstance(self.mil_layer, AttentionMIL):
mil_output = self.mil_layer({'features': features})
bag_features = mil_output['bag_features']
attention_weights = mil_output['attention_weights']
else:
features = features.view(batch_size * num_instances, -1)
bag_features = self.mil_layer(features.view(batch_size, num_instances, -1, 1, 1))
bag_features = bag_features.view(batch_size, -1)
attention_weights = None
# 分类
logits = self.classifier(bag_features)
return {
'logits': logits,
'attention_weights': attention_weights,
'bag_features': bag_features
}可视化和可解释性
注意力图可视化
def visualize_attention_map(wsi_path, model, attention_weights, patch_coordinates, save_path):
"""
在WSI上可视化注意力图
"""
import matplotlib.pyplot as plt
from matplotlib.patches import Rectangle
# 加载WSI
wsi = openslide.OpenSlide(wsi_path)
# 获取缩略图
thumbnail = wsi.get_thumbnail((wsi.dimensions[0] // 10, wsi.dimensions[1] // 10))
fig, ax = plt.subplots(1, 1, figsize=(15, 10))
ax.imshow(thumbnail)
# 绘制注意力权重
scale_factor = 10 # 缩略图缩放因子
for (x, y), weight in zip(patch_coordinates, attention_weights):
rect = Rectangle((x // scale_factor, y // scale_factor),
256 // scale_factor, 256 // scale_factor,
linewidth=1, edgecolor='red',
facecolor='red', alpha=weight.item())
ax.add_patch(rect)
ax.set_title('注意力图可视化')
ax.axis('off')
plt.tight_layout()
plt.savefig(save_path, dpi=150, bbox_inches='tight')
plt.close()🎯 医学影像目标检测技术
医学目标检测特征
与一般目标检测的差异
| 特征 | 一般目标检测 | 医学目标检测 |
|---|---|---|
| 目标 | 常见物体 | 病灶、结节、异常 |
| 尺寸变化 | 适中 | 很大(像素到全图) |
| 形状 | 规则 | 不规则、边界模糊 |
| 类别平衡 | 平衡 | 极度不平衡 |
| 标注成本 | 低 | 很高(专家标注) |
| 要求 | 速度 | 准确性和可解释性 |
特殊挑战
- 微小病灶:一些结节可能只有几个像素
- 低对比度:与周围组织密度相似
- 边界模糊:病灶边界不规则
- 类别不平衡:负样本远多于正样本
经典目标检测框架
Faster R-CNN适应
import torchvision
from torchvision.models.detection.faster_rcnn import FastRCNNPredictor
class MedicalFasterRCNN(nn.Module):
"""
为医学成像适配的Faster R-CNN
"""
def __init__(self, num_classes, pretrained=True):
super().__init__()
# 加载预训练Faster R-CNN
self.model = torchvision.models.detection.fasterrcnn_resnet50_fpn(pretrained=pretrained)
# 为灰度图像修改
self.model.backbone.body.conv1 = nn.Conv2d(1, 64, kernel_size=7, stride=2, padding=3, bias=False)
# 替换预测器
in_features = self.model.roi_heads.box_predictor.cls_score.in_features
self.model.roi_heads.box_predictor = FastRCNNPredictor(in_features, num_classes)
# 调整anchor以适应医学成像
self._adjust_anchors()
def _adjust_anchors(self):
"""
为医学病灶调整anchor框大小
"""
# 医学病灶通常较小
anchor_sizes = ((4, 8, 16), (8, 16, 32), (16, 32, 64))
aspect_ratios = ((0.5, 1.0, 2.0),) * len(anchor_sizes)
self.model.rpn.anchor_generator.sizes = anchor_sizes
self.model.rpn.anchor_generator.aspect_ratios = aspect_ratios
def forward(self, images, targets=None):
return self.model(images, targets)
def train_medical_detector(model, dataloader, optimizer, device, num_epochs):
"""
训练医学目标检测器
"""
model.train()
for epoch in range(num_epochs):
for images, targets in dataloader:
images = list(image.to(device) for image in images)
targets = [{k: v.to(device) for k, v in t.items()} for t in targets]
# 前向传播
loss_dict = model(images, targets)
losses = sum(loss for loss in loss_dict.values())
# 反向传播
optimizer.zero_grad()
losses.backward()
optimizer.step()
print(f"Epoch {epoch+1}/{num_epochs}, Loss: {losses.item():.4f}")YOLO适应
class MedicalYOLO(nn.Module):
"""
为医学成像适配的YOLO
"""
def __init__(self, num_classes, input_size=512, grid_size=16):
super().__init__()
self.input_size = input_size
self.grid_size = grid_size
self.num_classes = num_classes
self.num_boxes = 2 # 每个网格单元的box数量
# Backbone(简化Darknet)
self.backbone = self._create_backbone()
# 检测头
self.detection_head = self._create_detection_head()
def _create_backbone(self):
"""
创建backbone网络
"""
layers = []
# 第一个块
layers += [
nn.Conv2d(1, 32, 3, padding=1),
nn.BatchNorm2d(32),
nn.LeakyReLU(0.1),
nn.MaxPool2d(2),
nn.Conv2d(32, 64, 3, padding=1),
nn.BatchNorm2d(64),
nn.LeakyReLU(0.1),
nn.MaxPool2d(2),
nn.Conv2d(64, 128, 3, padding=1),
nn.BatchNorm2d(128),
nn.LeakyReLU(0.1),
nn.Conv2d(128, 64, 1),
nn.BatchNorm2d(64),
nn.LeakyReLU(0.1),
nn.Conv2d(64, 128, 3, padding=1),
nn.BatchNorm2d(128),
nn.LeakyReLU(0.1),
nn.MaxPool2d(2),
]
return nn.Sequential(*layers)
def _create_detection_head(self):
"""
创建检测头
"""
feature_size = 128 * (self.input_size // 32) ** 2
grid_total = self.grid_size ** 2
return nn.Sequential(
nn.Linear(feature_size, 1024),
nn.LeakyReLU(0.1),
nn.Dropout(0.5),
nn.Linear(1024, grid_total * self.num_boxes * (5 + self.num_classes)),
nn.Sigmoid()
)
def forward(self, x):
features = self.backbone(x)
batch_size = features.size(0)
features = features.view(batch_size, -1)
detections = self.detection_head(features)
detections = detections.view(
batch_size, self.grid_size, self.grid_size,
self.num_boxes, 5 + self.num_classes
)
return detections损失函数设计
目标检测的Focal Loss
class FocalLossForDetection(nn.Module):
"""
医学目标检测的Focal Loss
"""
def __init__(self, alpha=0.25, gamma=2.0):
super().__init__()
self.alpha = alpha
self.gamma = gamma
def forward(self, predictions, targets):
"""
Args:
predictions: (batch_size, grid_size, grid_size, num_boxes, 5 + num_classes)
targets: (batch_size, grid_size, grid_size, num_boxes, 5 + num_classes)
"""
# 目标性损失
pred_obj = predictions[..., 4]
target_obj = targets[..., 4]
focal_loss = self.alpha * (1 - pred_obj) ** self.gamma * \
F.binary_cross_entropy(pred_obj, target_obj, reduction='none')
# 类别损失(仅对目标单元)
pred_class = predictions[..., 5:]
target_class = targets[..., 5:]
class_loss = F.binary_cross_entropy(pred_class, target_class, reduction='none')
# 仅在有目标的地方应用
class_loss = class_loss * target_obj.unsqueeze(-1)
# 坐标损失(仅对目标单元)
pred_coord = predictions[..., :4]
target_coord = targets[..., :4]
coord_loss = F.mse_loss(pred_coord, target_coord, reduction='none')
coord_loss = coord_loss.sum(dim=-1) * target_obj
# 总损失
total_loss = focal_loss + class_loss.sum(dim=-1) + coord_loss
return total_loss.mean()医学专用优化策略
难负样本挖掘
class HardNegativeMiner:
"""
医学目标检测的难负样本挖掘
"""
def __init__(self, negative_ratio=3):
self.negative_ratio = negative_ratio
def mine_hard_negatives(self, predictions, targets):
"""
挖掘难负样本
"""
# 计算每个预测框和真实框的IoU
ious = self._calculate_ious(predictions, targets)
# 识别负样本(IoU < 阈值)
negative_mask = ious < 0.3
negative_indices = torch.where(negative_mask)
if len(negative_indices[0]) == 0:
return None
# 计算负样本的置信度分数
negative_confidences = predictions[negative_indices][:, 4]
# 选择难负样本(置信度最高的负样本)
num_negatives = min(len(negative_confidences),
self.negative_ratio * len(torch.where(ious >= 0.5)[0]))
_, hard_indices = torch.topk(negative_confidences, num_negatives)
return negative_indices[0][hard_indices]
def _calculate_ious(self, predictions, targets):
"""
计算预测框和目标框之间的IoU
"""
# 简化的IoU计算
pred_boxes = predictions[:, :4]
target_boxes = targets[:, :4]
# 计算交集
inter_x1 = torch.max(pred_boxes[:, 0], target_boxes[:, 0])
inter_y1 = torch.max(pred_boxes[:, 1], target_boxes[:, 1])
inter_x2 = torch.min(pred_boxes[:, 2], target_boxes[:, 2])
inter_y2 = torch.min(pred_boxes[:, 3], target_boxes[:, 3])
inter_area = torch.clamp(inter_x2 - inter_x1, min=0) * \
torch.clamp(inter_y2 - inter_y1, min=0)
# 计算并集
pred_area = (pred_boxes[:, 2] - pred_boxes[:, 0]) * (pred_boxes[:, 3] - pred_boxes[:, 1])
target_area = (target_boxes[:, 2] - target_boxes[:, 0]) * (target_boxes[:, 3] - target_boxes[:, 1])
union_area = pred_area + target_area - inter_area
return inter_area / (union_area + 1e-8)📊 性能对比与实践指南
评估指标
分类指标
def calculate_medical_classification_metrics(y_true, y_pred, y_prob=None):
"""
计算医学分类评估指标
"""
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score
from sklearn.metrics import roc_auc_score, confusion_matrix, classification_report
metrics = {}
# 基础指标
metrics['accuracy'] = accuracy_score(y_true, y_pred)
metrics['precision'] = precision_score(y_true, y_pred, average='weighted')
metrics['recall'] = recall_score(y_true, y_pred, average='weighted')
metrics['f1_score'] = f1_score(y_true, y_pred, average='weighted')
# 医学专用指标
metrics['sensitivity'] = recall_score(y_true, y_pred, pos_label=1) # 真阳性率
metrics['specificity'] = recall_score(y_true, y_pred, pos_label=0,
average='binary') # 真阴性率
# AUC-ROC
if y_prob is not None:
metrics['auc_roc'] = roc_auc_score(y_true, y_prob[:, 1])
# 混淆矩阵
cm = confusion_matrix(y_true, y_pred)
metrics['confusion_matrix'] = cm
# 详细报告
metrics['classification_report'] = classification_report(y_true, y_pred)
return metrics检测指标
def calculate_detection_metrics(predictions, targets, iou_threshold=0.5):
"""
计算目标检测指标
"""
detections = []
ground_truths = []
for pred, target in zip(predictions, targets):
# 转换为标准格式
det_boxes = pred['boxes'].cpu().numpy()
det_scores = pred['scores'].cpu().numpy()
det_labels = pred['labels'].cpu().numpy()
gt_boxes = target['boxes'].cpu().numpy()
gt_labels = target['labels'].cpu().numpy()
# 计算IoU矩阵
iou_matrix = calculate_iou_matrix(det_boxes, gt_boxes)
# 匹配预测与真实值
matches = match_detections(iou_matrix, iou_threshold)
detections.extend(det_boxes)
ground_truths.extend(gt_boxes)
# 计算mAP
mAP = calculate_map(detections, ground_truths, iou_threshold)
return {
'mAP': mAP,
'total_predictions': len(detections),
'total_ground_truths': len(ground_truths)
}
def calculate_map(detections, ground_truths, iou_threshold=0.5):
"""
计算平均精度均值(mAP)
"""
# 按置信度排序
detections.sort(key=lambda x: x['score'], reverse=True)
# 计算精确度和召回率
tp = 0
fp = 0
fn = len(ground_truths)
precisions = []
recalls = []
for det in detections:
# 检查是否为真阳性
is_tp = False
for gt in ground_truths:
if calculate_iou(det['box'], gt['box']) > iou_threshold:
tp += 1
fn -= 1
is_tp = True
break
if not is_tp:
fp += 1
precision = tp / (tp + fp) if (tp + fp) > 0 else 0
recall = tp / (tp + fn) if (tp + fn) > 0 else 0
precisions.append(precision)
recalls.append(recall)
# 计算AP
ap = calculate_ap_from_pr(precisions, recalls)
return ap模型选择指南
任务驱动的模型选择
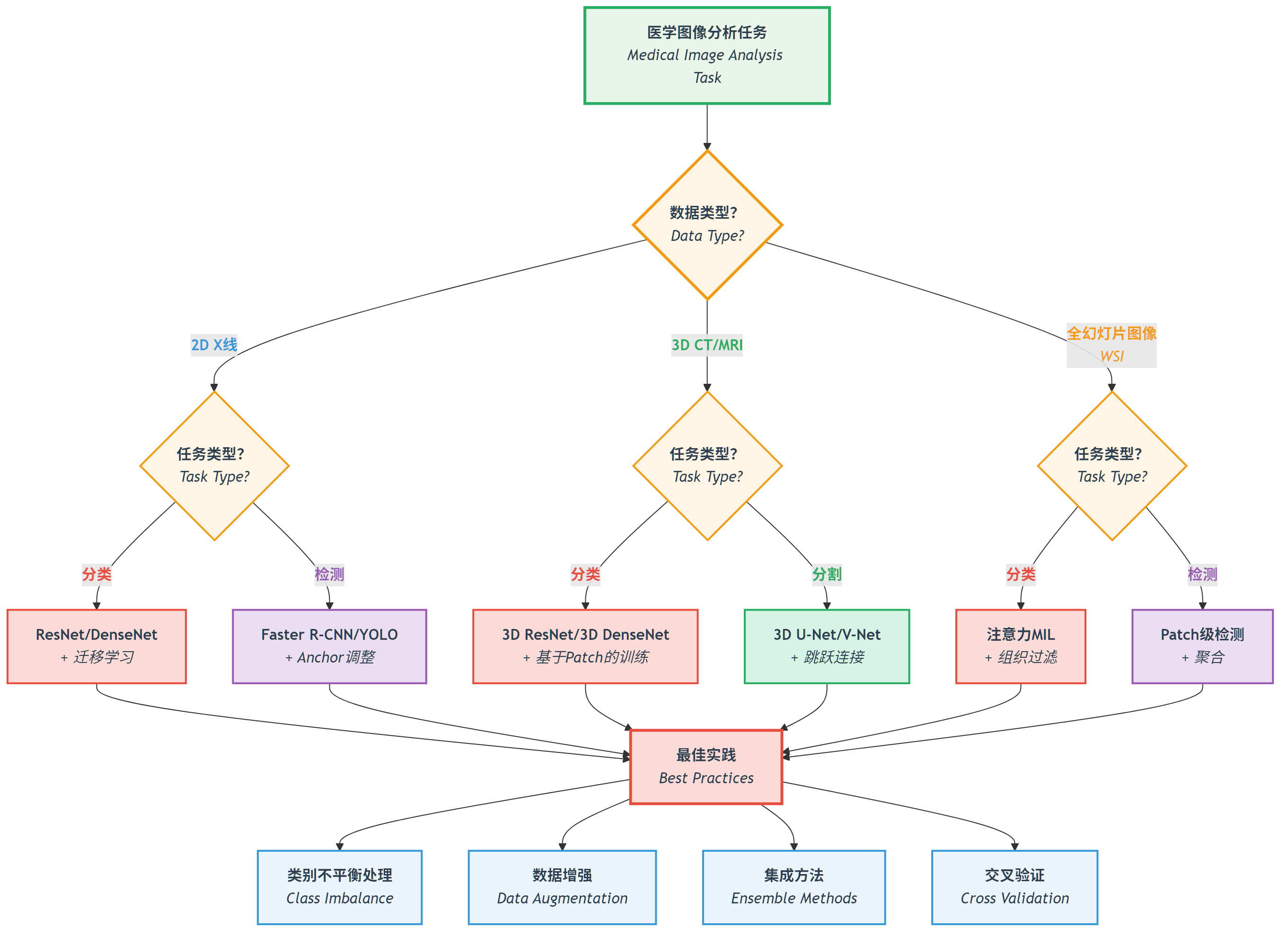 *图:根据医学图像的数据类型(2D X线、3D CT/MRI、WSI全幻灯片图像)和任务类型选择合适的深度学习模型 *
*图:根据医学图像的数据类型(2D X线、3D CT/MRI、WSI全幻灯片图像)和任务类型选择合适的深度学习模型 *
📖 查看原始Mermaid代码
性能比较
| 模型 | 数据类型 | 任务 | mAP/准确率 | 内存使用 | 训练时间 | 临床适用性 |
|---|---|---|---|---|---|---|
| ResNet50 | 2D X线 | 分类 | 0.85-0.92 | 2GB | 中等 | ⭐⭐⭐⭐⭐ |
| DenseNet121 | 2D X线 | 分类 | 0.87-0.94 | 2.5GB | 中等 | ⭐⭐⭐⭐⭐ |
| 3D ResNet | 3D CT/MRI | 分类 | 0.82-0.89 | 8GB | 高 | ⭐⭐⭐⭐⭐ |
| Faster R-CNN | 2D X线 | 检测 | 0.78-0.85 | 4GB | 高 | ⭐⭐⭐⭐⭐ |
| YOLOv5 | 2D X线 | 检测 | 0.75-0.82 | 1.5GB | 低 | ⭐⭐⭐⭐⭐ |
| 注意力MIL | WSI | 分类 | 0.80-0.88 | 6GB | 非常高 | ⭐⭐⭐⭐⭐ |
🎯 技术要点与发展趋势
1. 分类技术
- 2D CNN用于X线:基于ResNet、DenseNet的迁移学习
- 3D CNN用于体积数据:3D ResNet、内存优化策略
- 数据不平衡处理:Focal Loss、平衡采样
2. 检测策略
- 经典框架:Faster R-CNN、YOLO医学适应
- 医学专用:难负样本挖掘、anchor调整
- 评估指标:mAP、IoU、临床指标
3. 全幻灯片图像分析
- MIL框架:注意力机制、实例级学习
- 内存效率:基于Patch的处理、缓存策略
- 可解释性:注意力图可视化
4. 最佳实践
- 临床要求:准确率第一,速度第二
- 数据质量:高质量标注、多中心验证
- 法规合规:模型可解释性、决策支持
5. 未来趋势
- 多模态融合:结合成像和临床数据的综合分析
- 弱监督学习:减少标注需求
- 联邦学习:多中心合作、隐私保护
🔗 本章节相关的典型医学数据集和论文网址与开源库
Details
数据集
| 数据集 | 用途 | 官方网址 | 许可证 | 备注 |
|---|---|---|---|---|
| NIH ChestX-ray14 | 胸部X光分类检测 | https://nihcc.app.box.com/v/ChestX-ray14 | 公开 | 包含14种胸部疾病标签 |
| CheXpert | 胸部X光分类 | https://stanfordmlgroup.github.io/competitions/chexpert/ | CC-BY 4.0 | Stanford标准数据集,含5种异常标签 |
| MIMIC-CXR | 胸部X光多标签分类 | https://physionet.org/content/mimic-cxr-jpg/2.0.0/ | MIT许可 | 波士顿儿童医院真实临床数据 |
| PadChest | 胸部X光+临床数据 | https://bimcv.cipf.es/bimcv-projects/padchest/ | CC BY 4.0 | 包含10万张X光片,含临床报告 |
| DeepLesion | 病灶检测数据集 | https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=53683303 | 公开 | 包含各种体部病灶的标注数据 |
| MedicalDecathlon | 多器官分类分割 | https://medicaldecathlon.com/ | CC BY-SA 4.0 | 10个器官的CT/MRI数据集 |
| ChestX-Ray8 | 胸部疾病分类 | https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia | 公开 | 包含肺炎、正常X光片 |
| ISIC Archive | 皮肤病变分类 | https://www.isic-archive.com/#!/topWithHeader/onlyHeaderTop/gallery | 公开 | 皮肤镜图像分类基准 |
论文
| 论文标题 | 关键字段 | 来源 | 备注 |
|---|---|---|---|
| CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning | 胸部X光肺炎检测 | arXiv:1711.05225 | Stanford大学,使用121层DenseNet |
| Focal Loss for Dense Object Detection | Focal Loss损失函数 | arXiv:1708.02002 | 解决类别不平衡的经典损失函数论文 |
开源库
| 库名称 | 功能 | GitHub/官网 | 用途 |
|---|---|---|---|
| MONAI | 医学影像深度学习框架 | https://monai.io/ | 专为医学影像设计的PyTorch库,包含分类、检测、分割工具 |
| TorchIO | 医学图像变换库 | https://torchio.readthedocs.io/ | 支持多种医学图像格式和增强变换 |
| deepmedic | 3D医学图像分类 | https://github.com/DeepMedic/deepmedic | 高性能3D医学图像分类框架,特别适合脑部图像 |
| Grad-CAM++ | 可解释性可视化 | https://github.com/jacobgil/grad-cam-plus-plus | 医学图像分类的注意力可视化工具 |
| medcam | 医学CAM实现 | https://github.com/rigetti/medcam | 针对医学图像的可解释性CAM工具 |
| clEsperanto | GPU加速医学图像处理 | https://github.com/CI-BinaryDisease/clEsperanto | 基于GPU的医学图像处理和分类加速 |
| pydicom | DICOM文件处理 | https://pydicom.github.io/pydicom-sphinx/ | 医学DICOM文件的读取和处理 |
| SimpleITK | 医学图像处理 | https://simpleitk.org/ | 跨平台的医学图像处理库,支持多种格式 |
| OpenSlide | 全幻灯片图像处理 | https://openslide.org/ | 用于处理全幻灯片图像的高性能库 |
🚀 下一步学习
现在你已经掌握了医学图像分类和检测的核心技术。在下一节(5.4 图像增强和恢复)中,我们将探讨如何通过先进的数据增强技术克服数据稀缺挑战,并恢复丢失的图像信息。